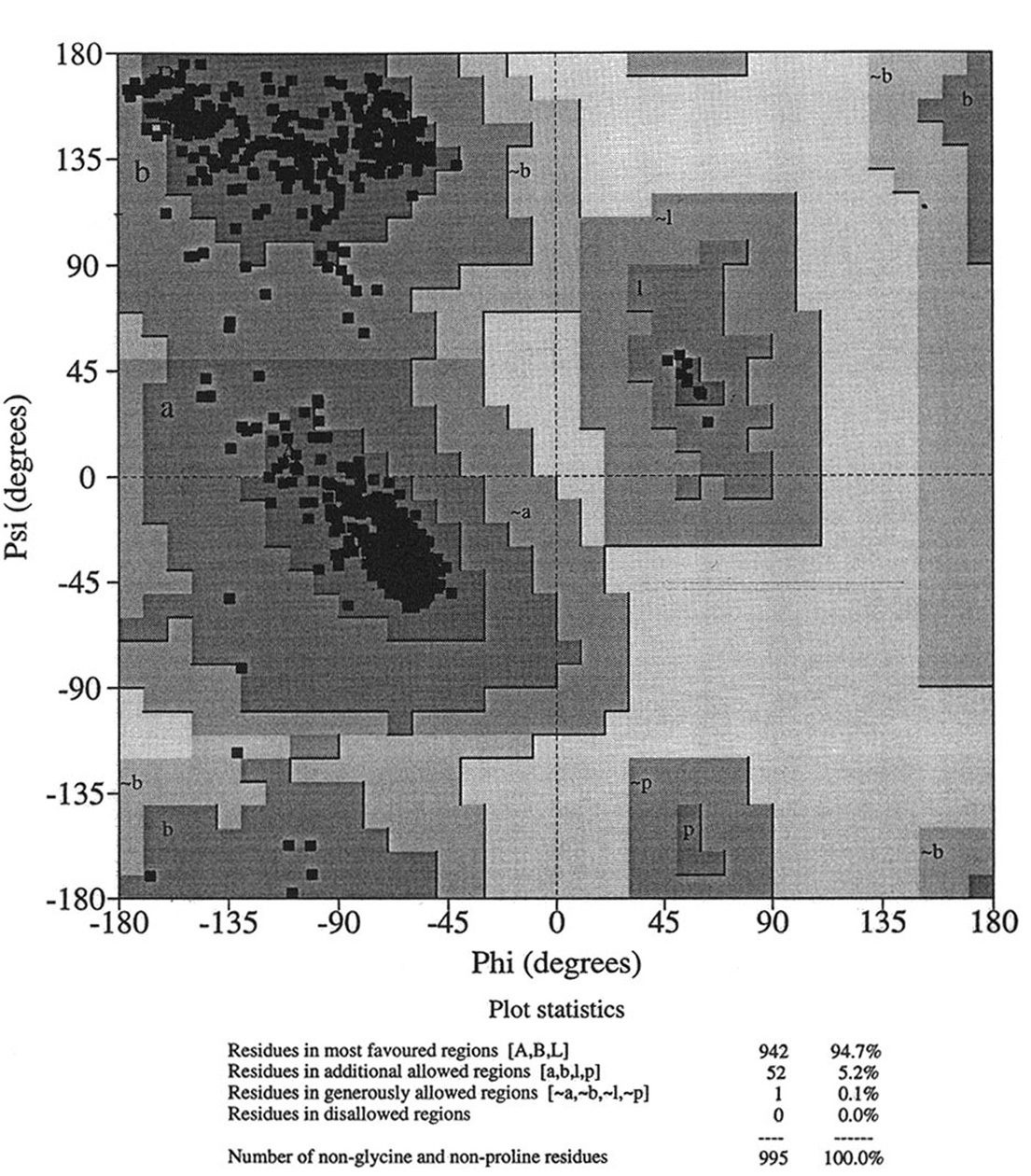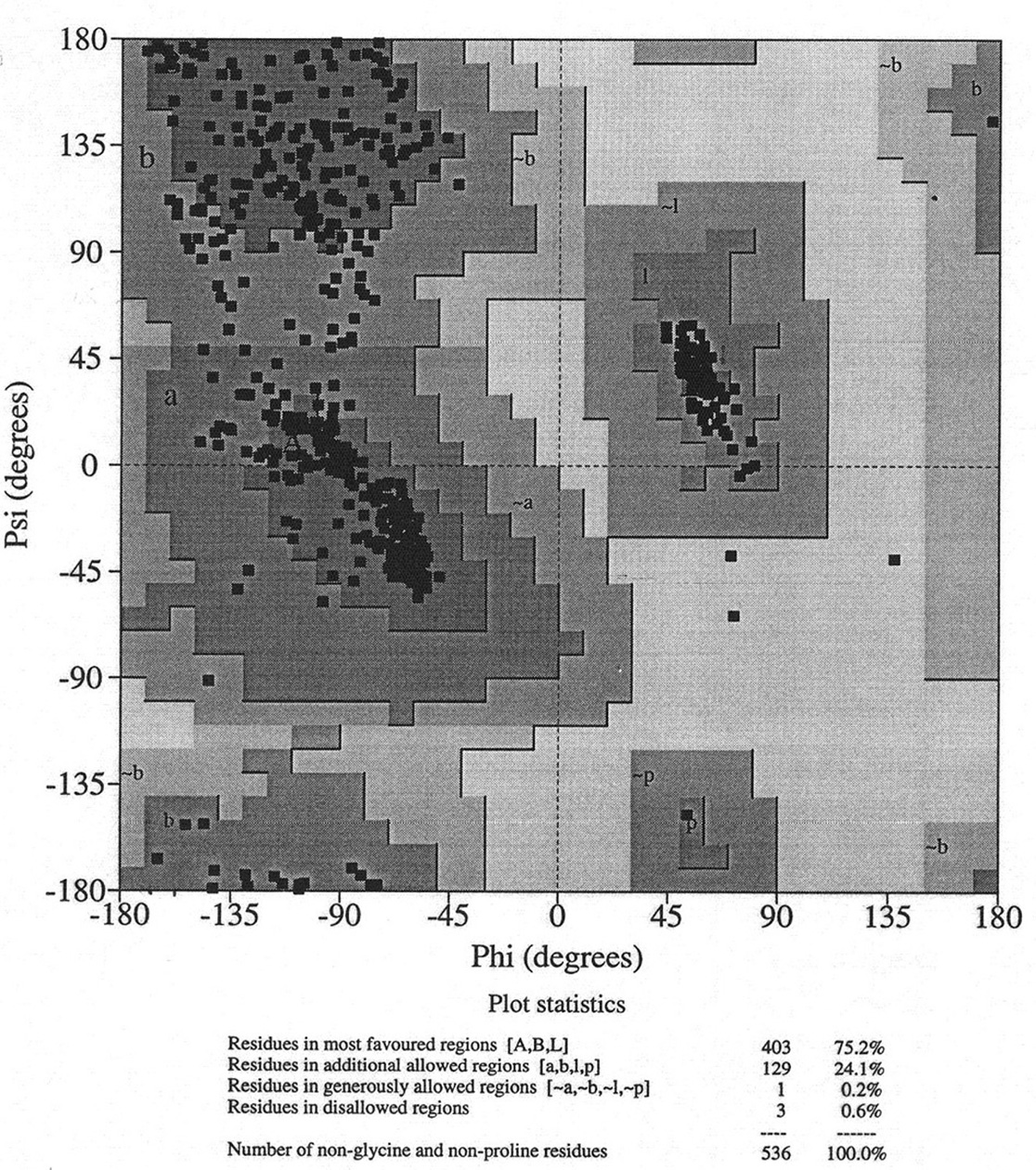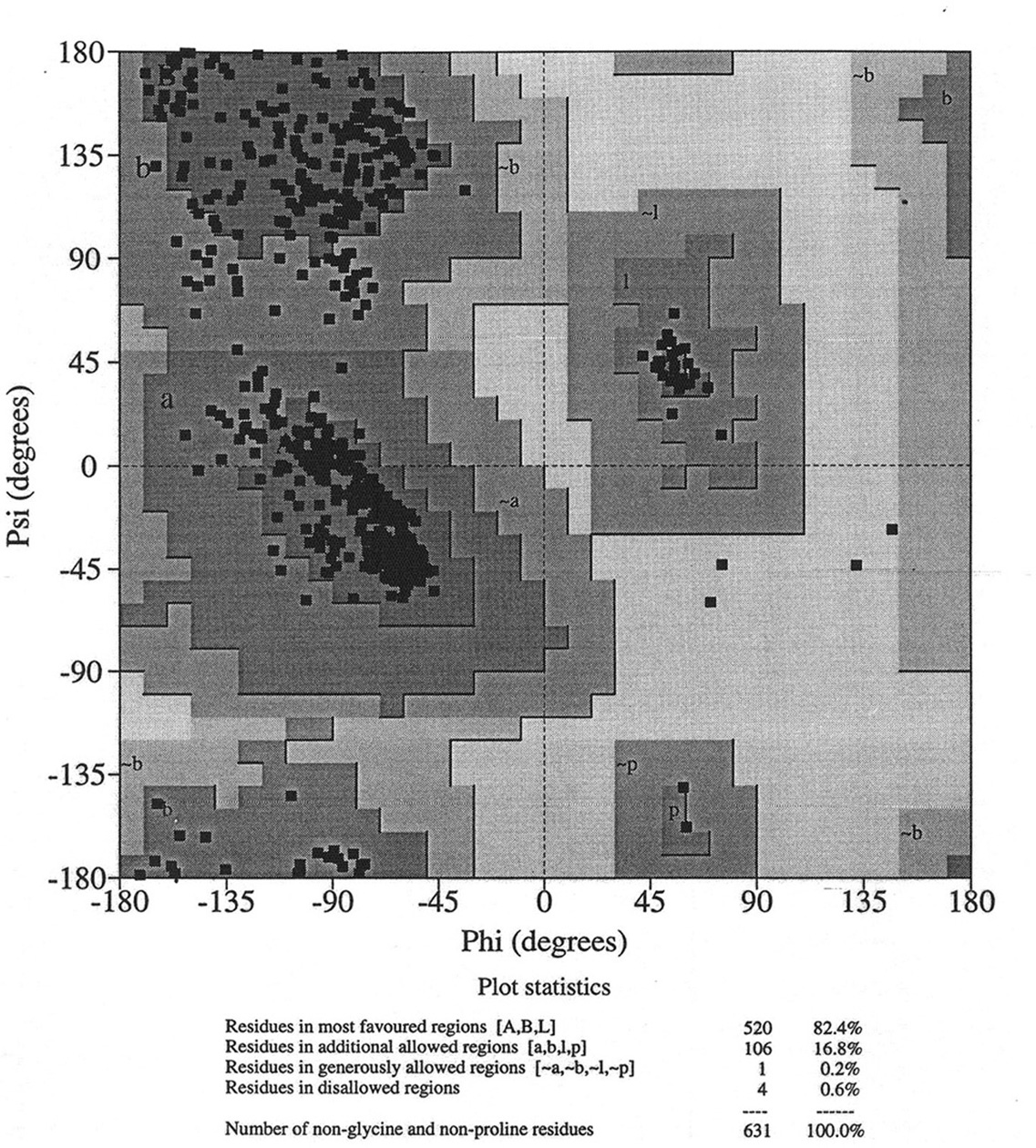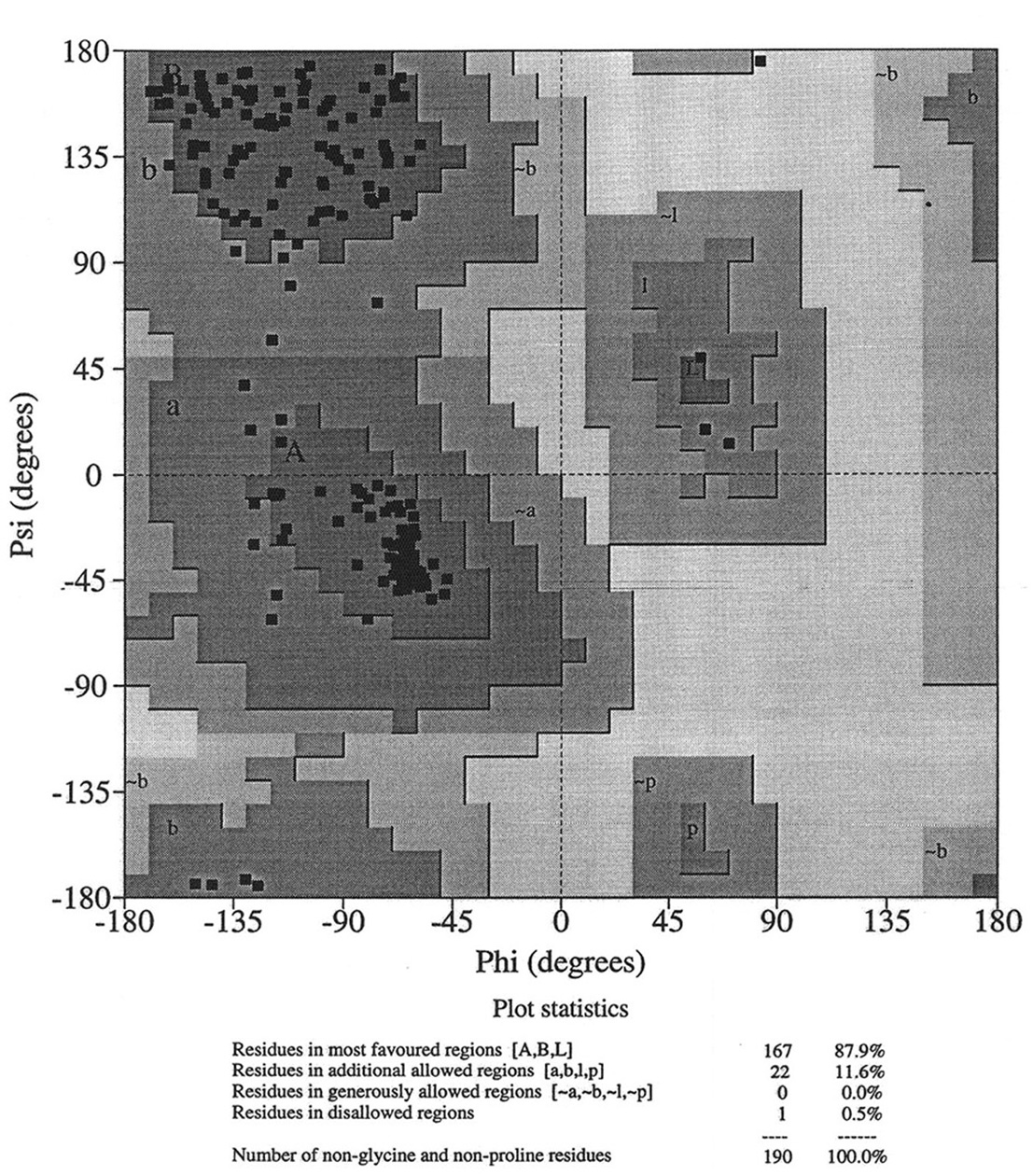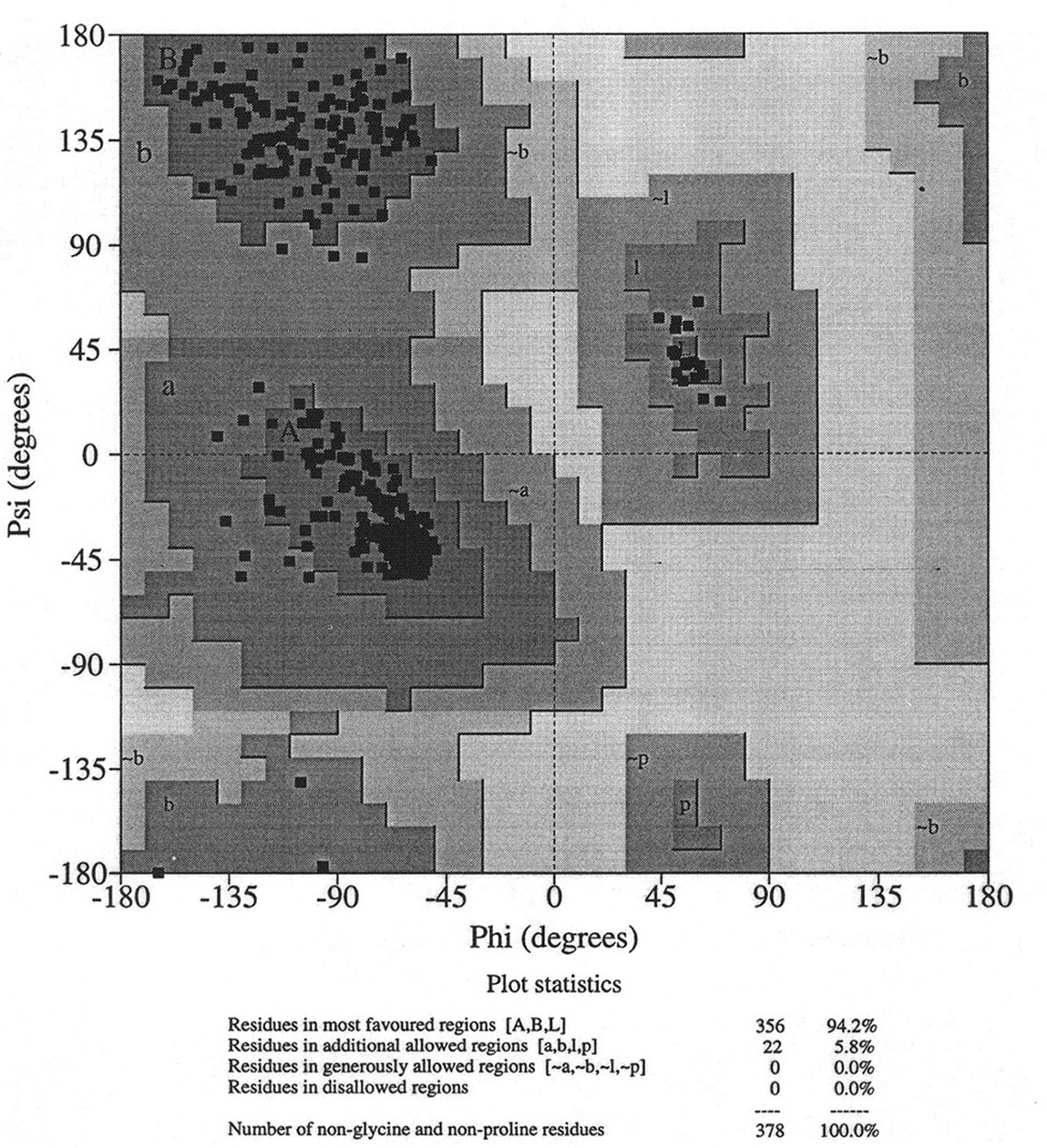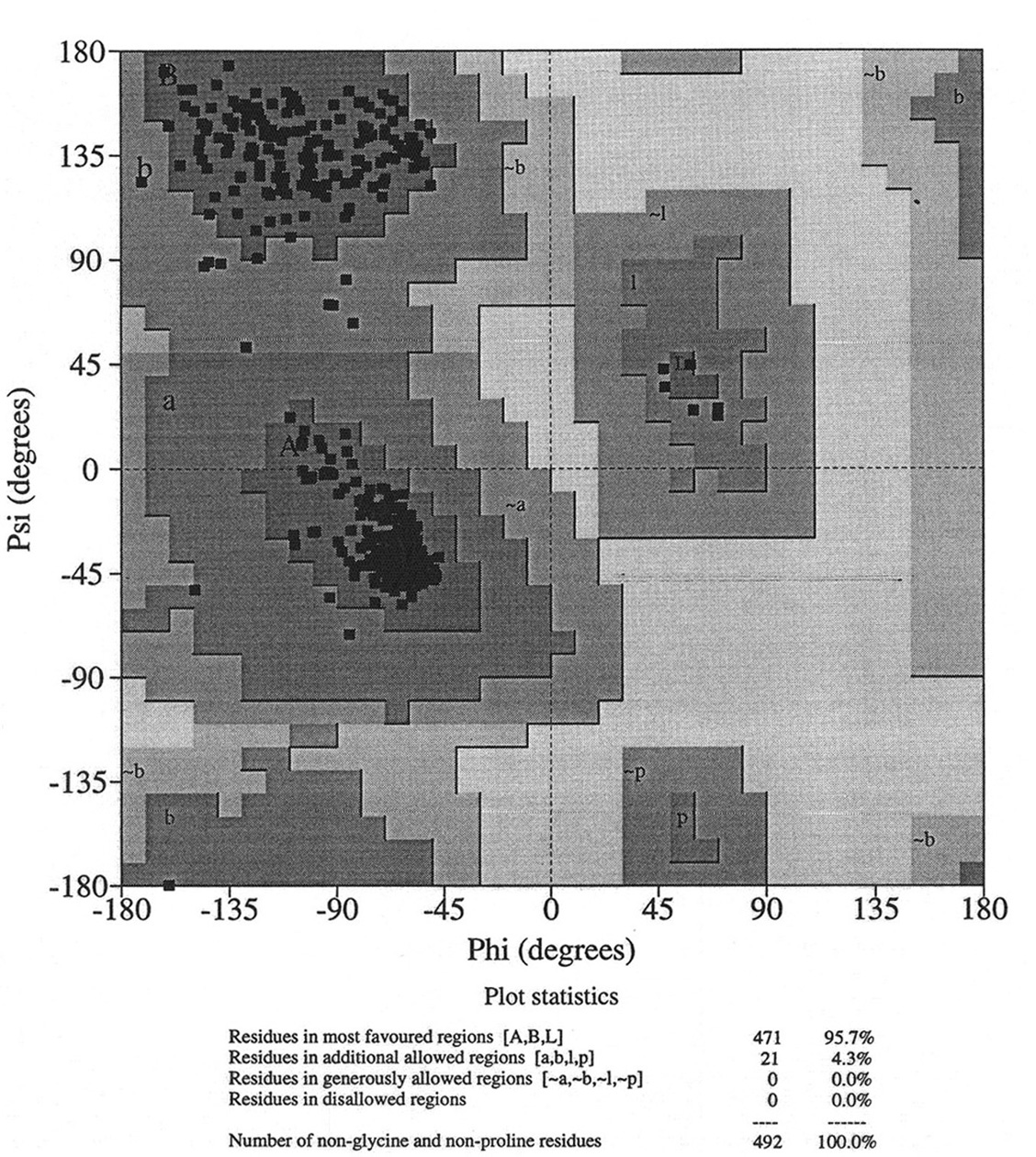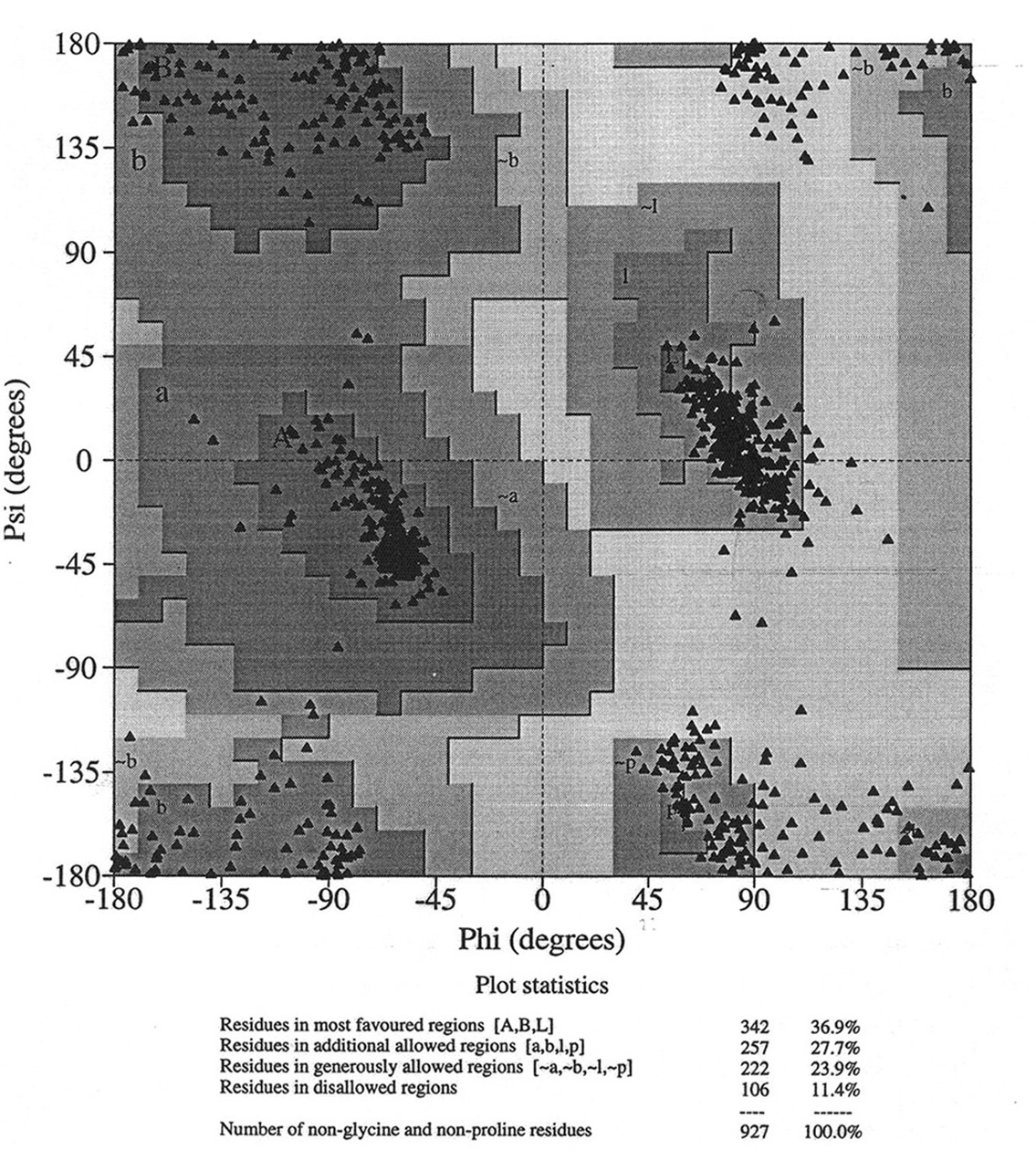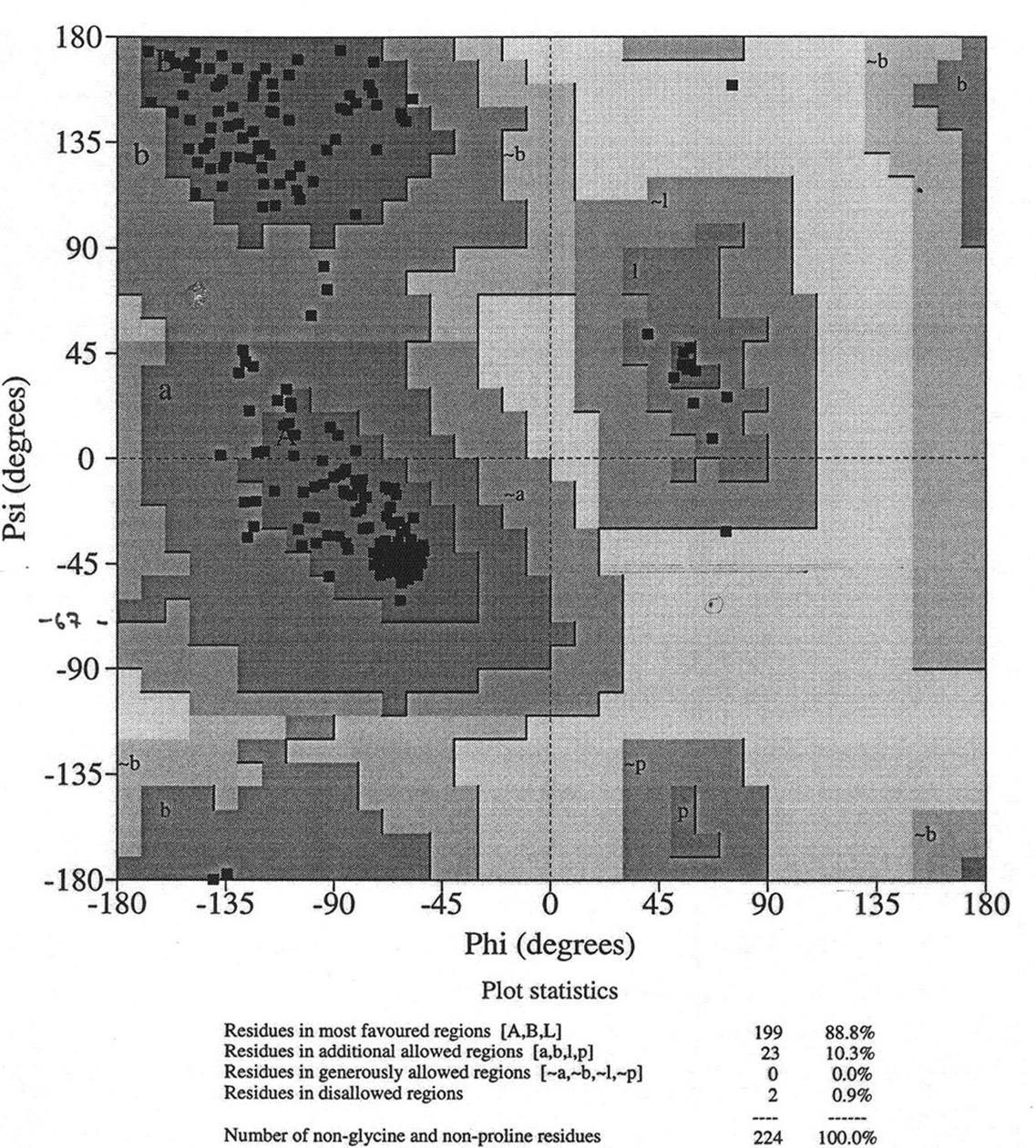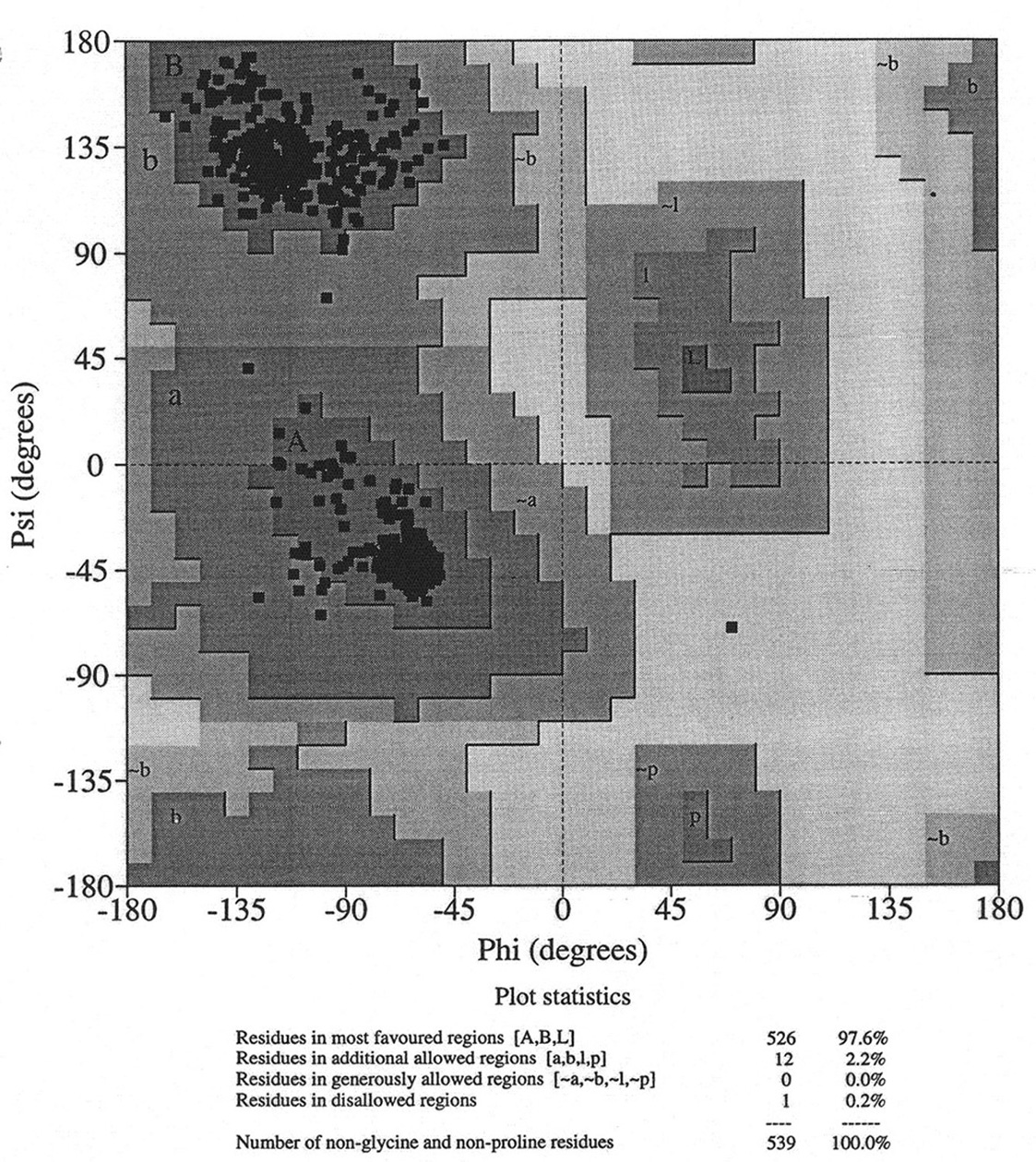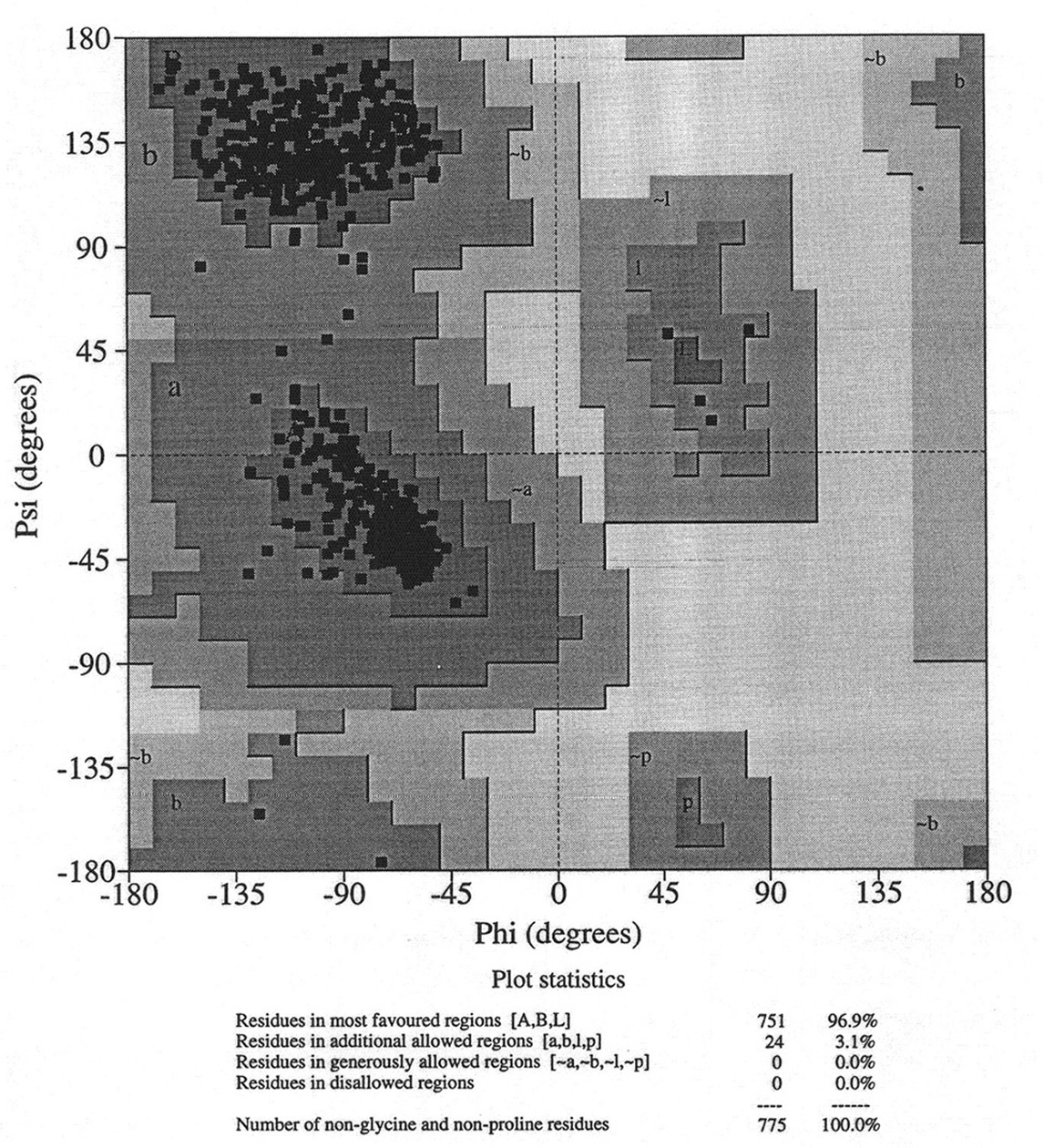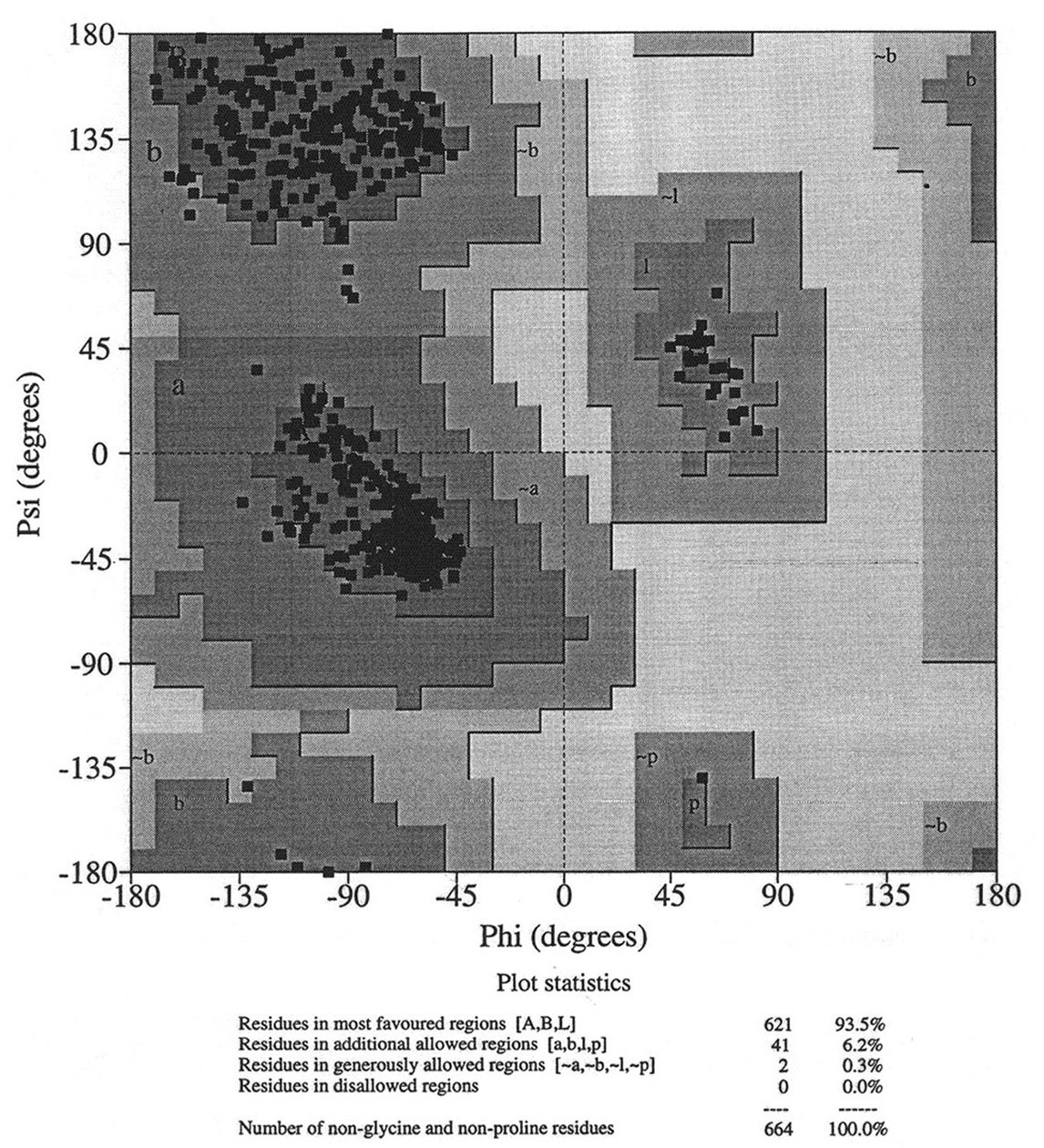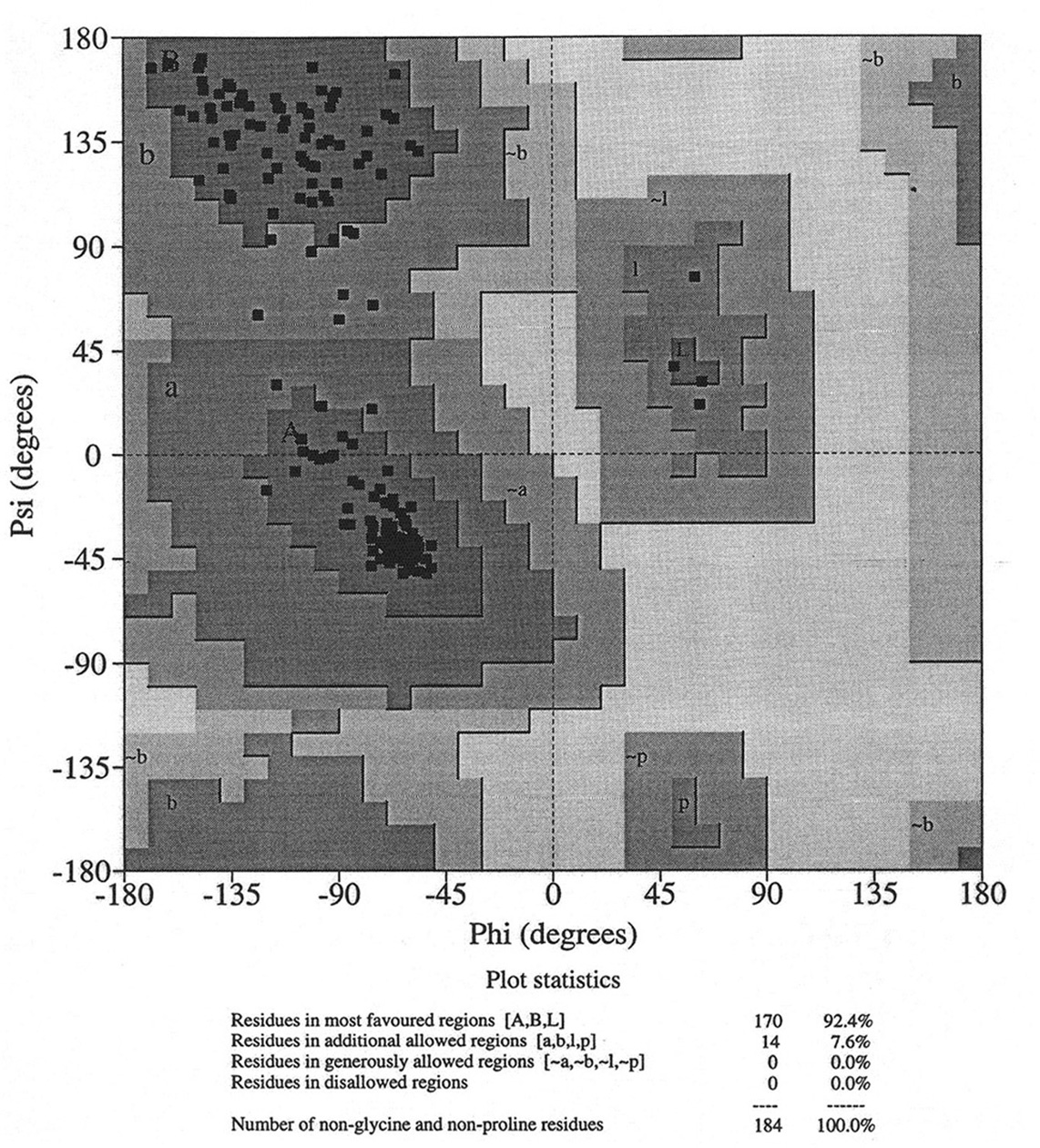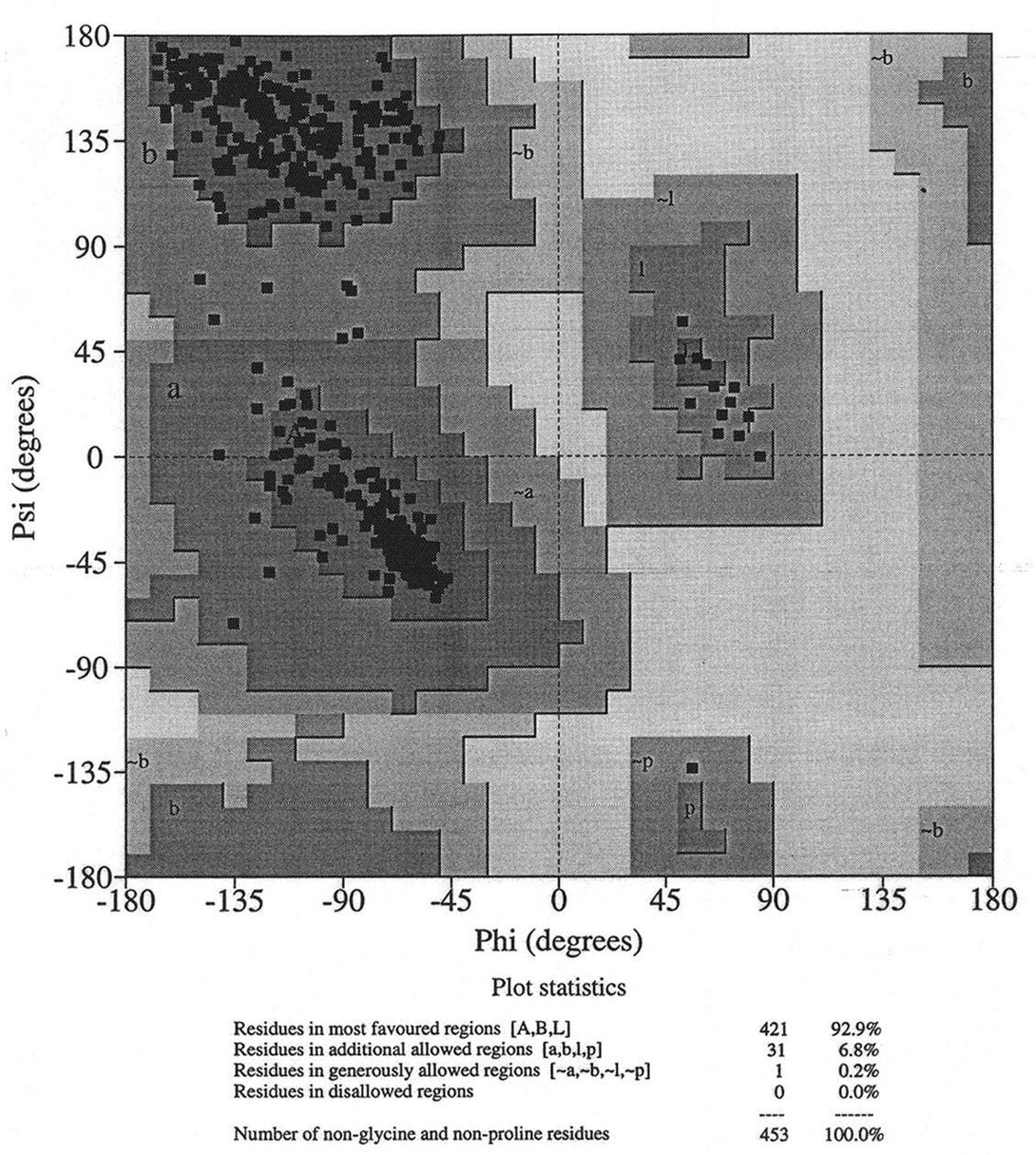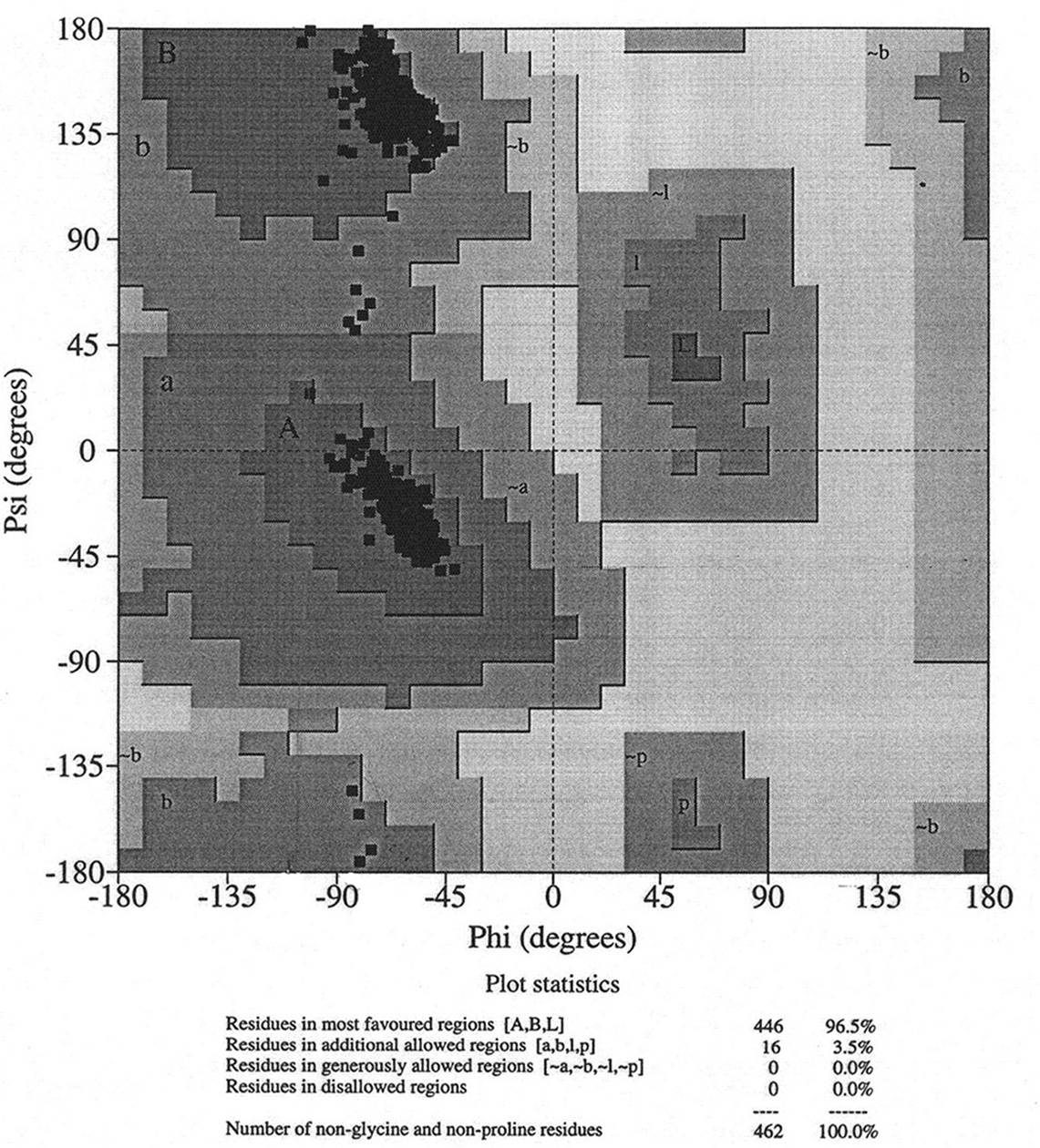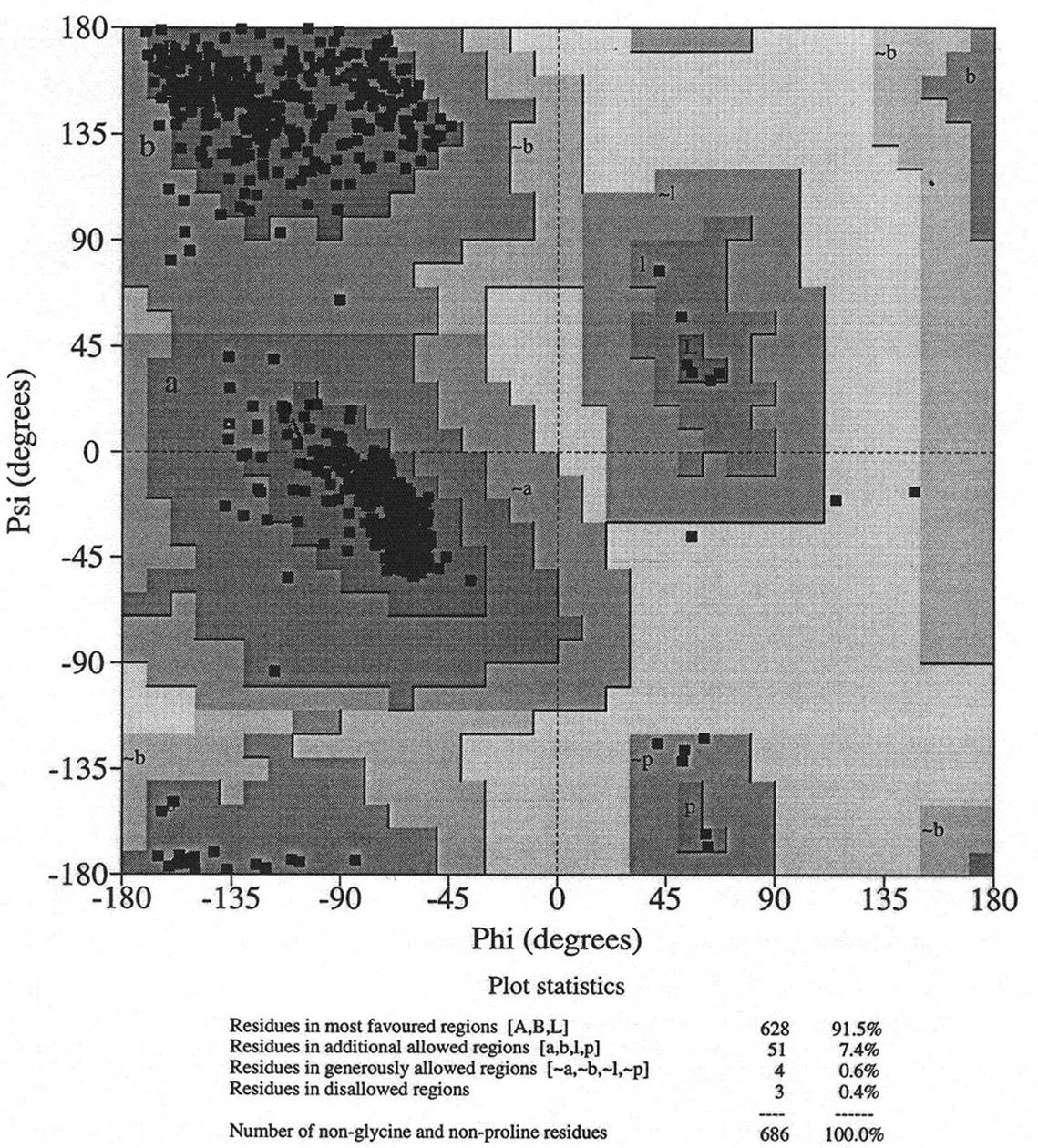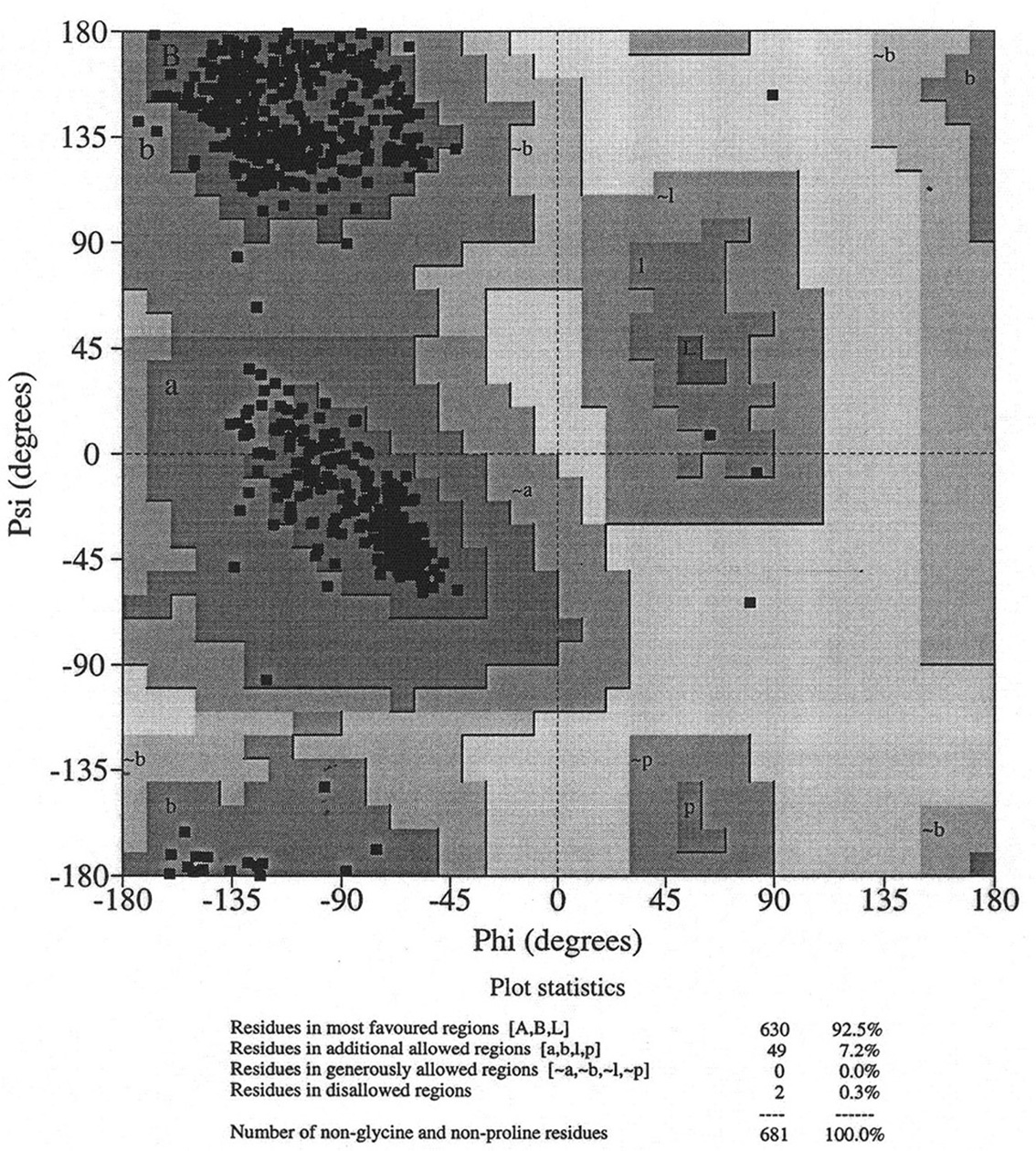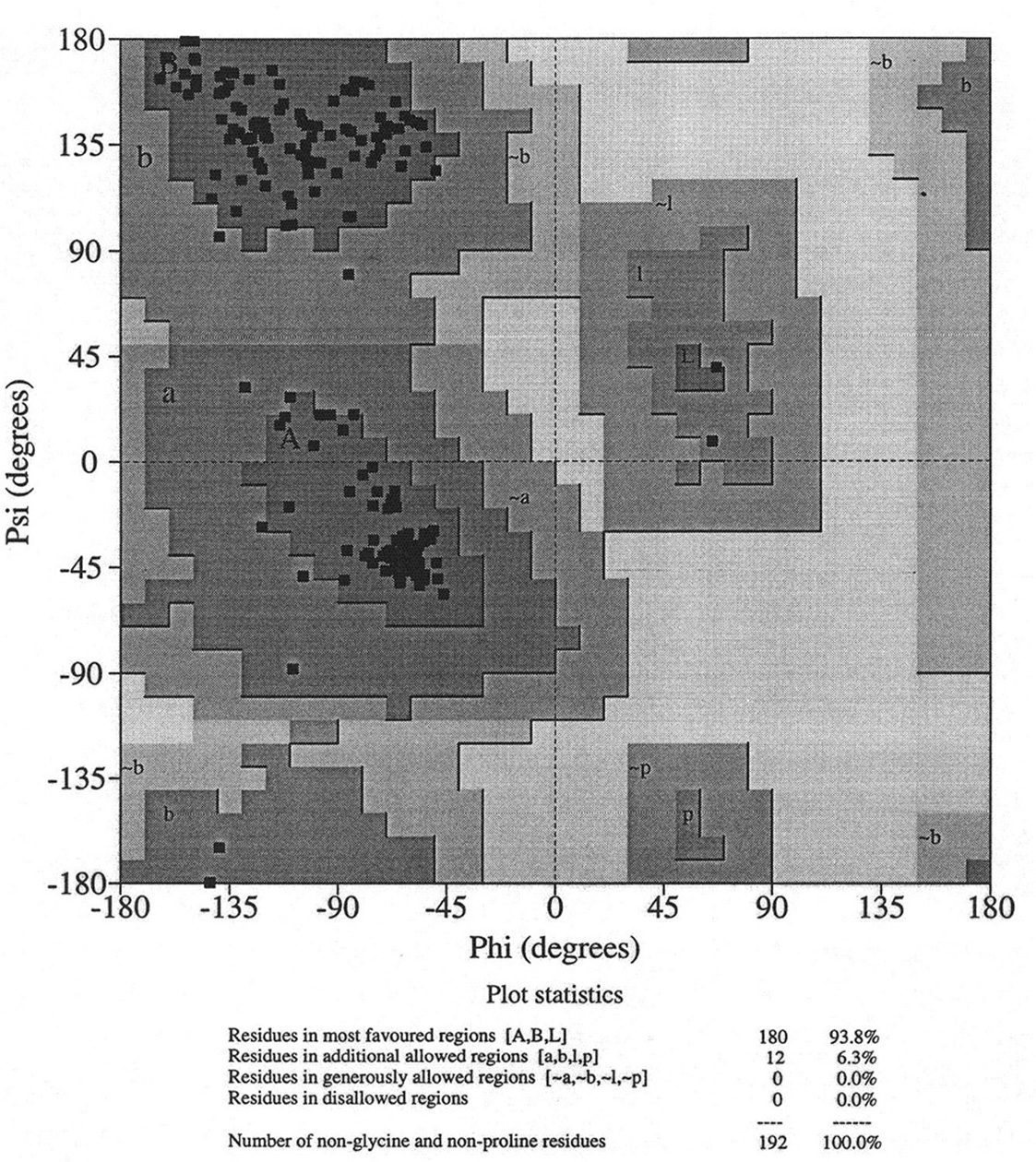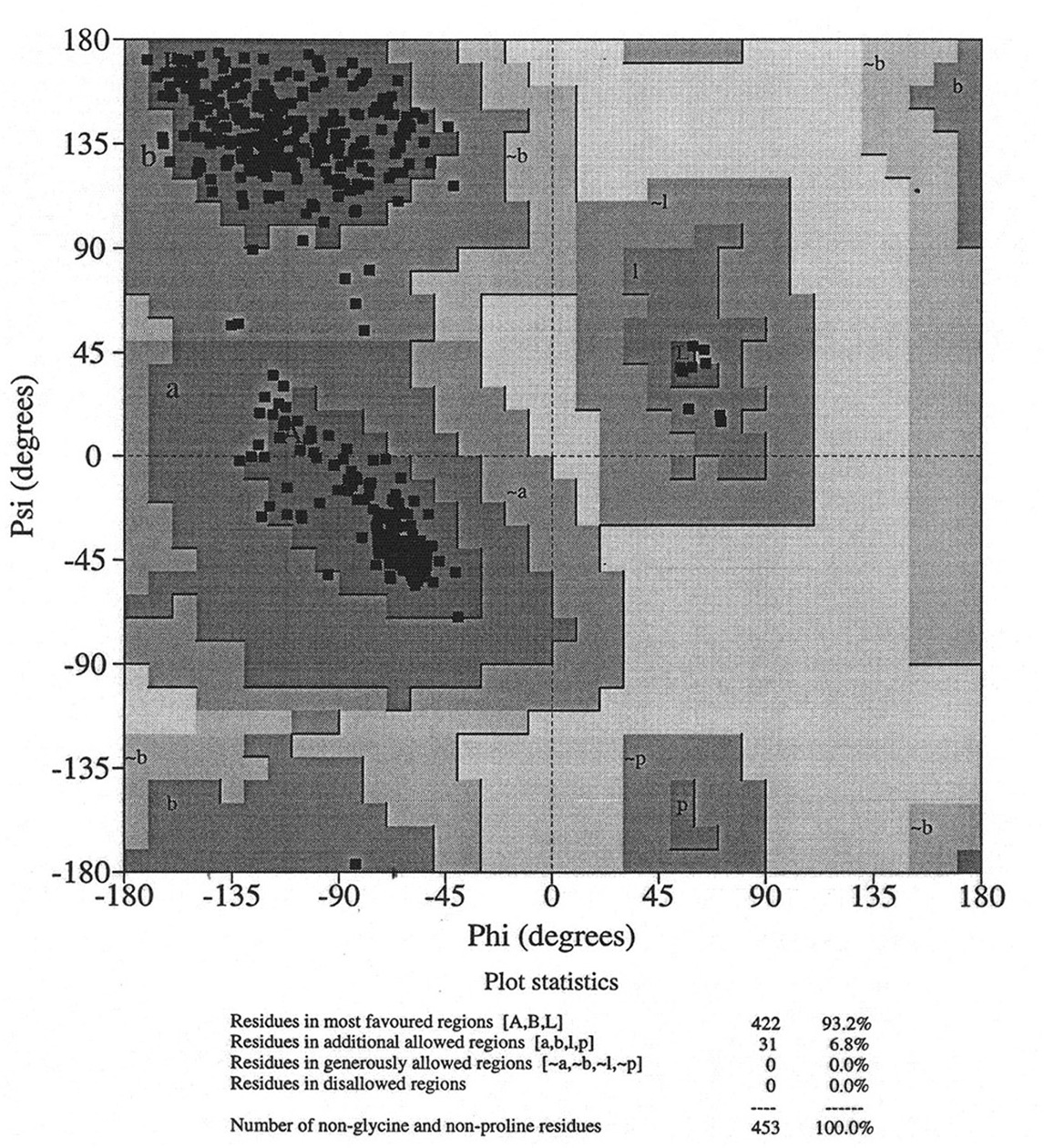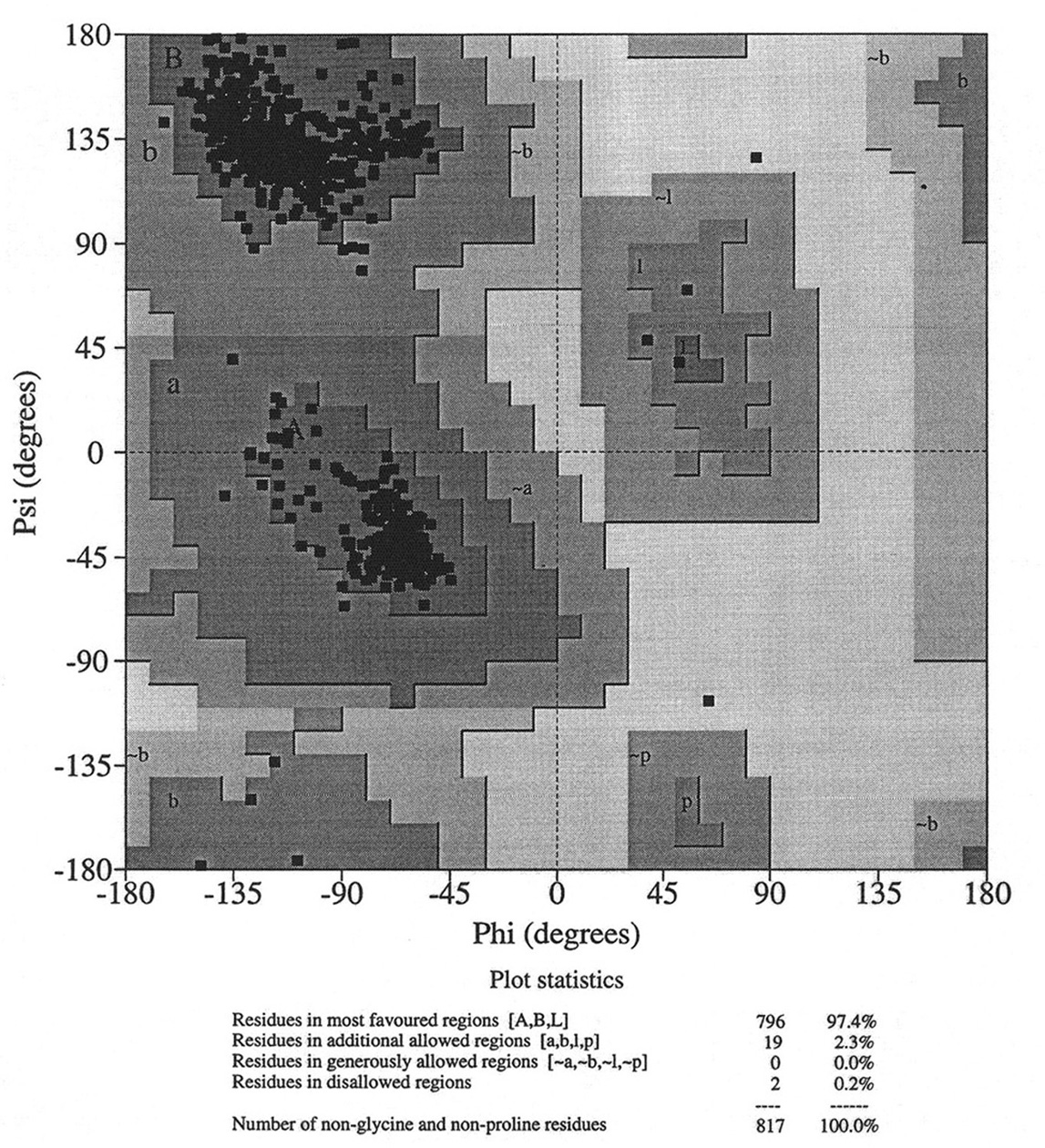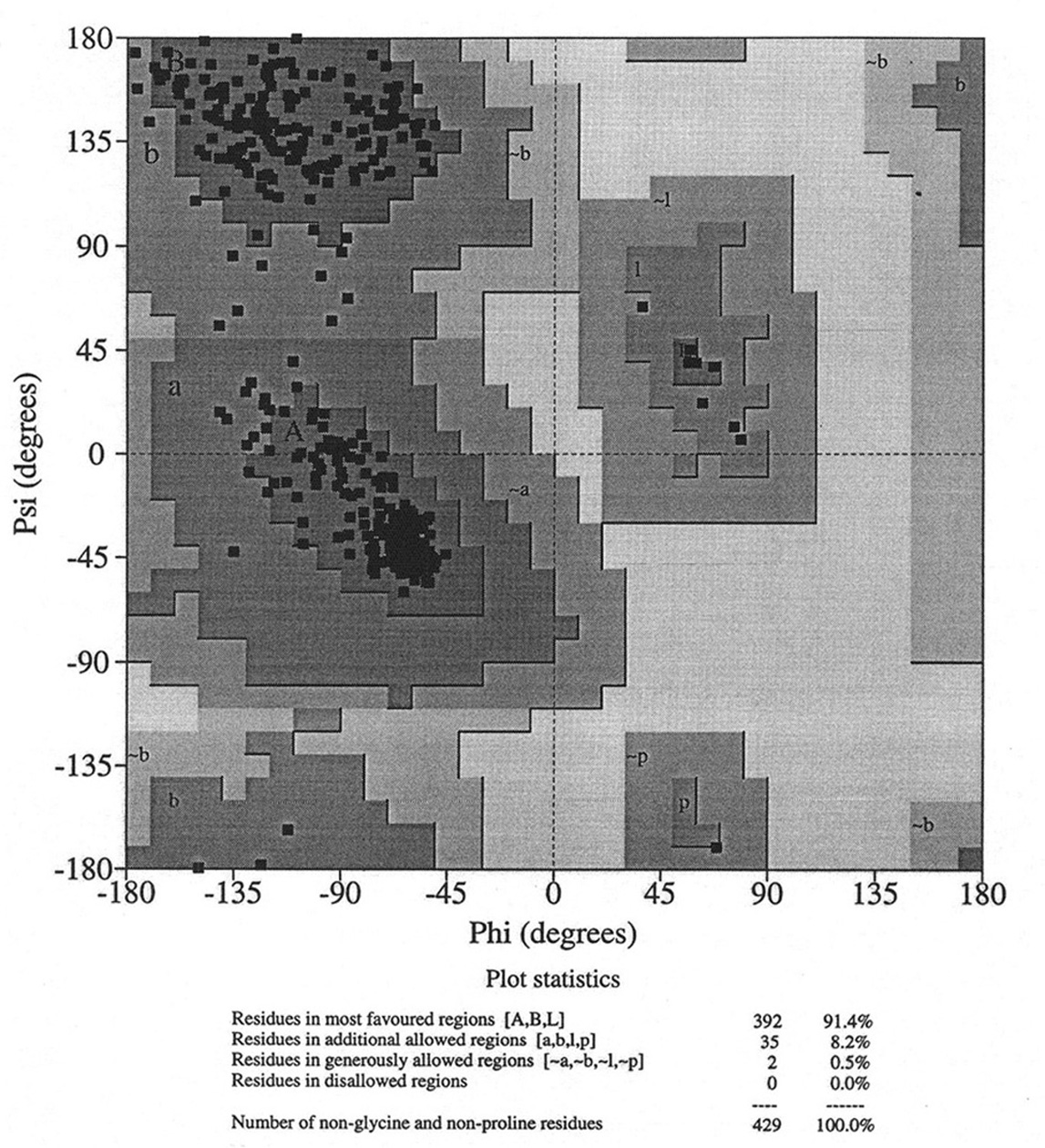
Database of 70 structures assembled by Andy Karplus [Karplus, P.A. (1996) Experimentally observed conformation-dependent geometry and hidden strain in proteins. Protein. Sci. 5, 1406, 14020]
Kuszewski, J., Gronenborn, A.M. & Clore, G.M. (1996) Improving the quality of NMR and crystallographic protein structures by means of a conformational database potential derived from structure databases. Protein Science 5, 1067-1080. pubmed PDF